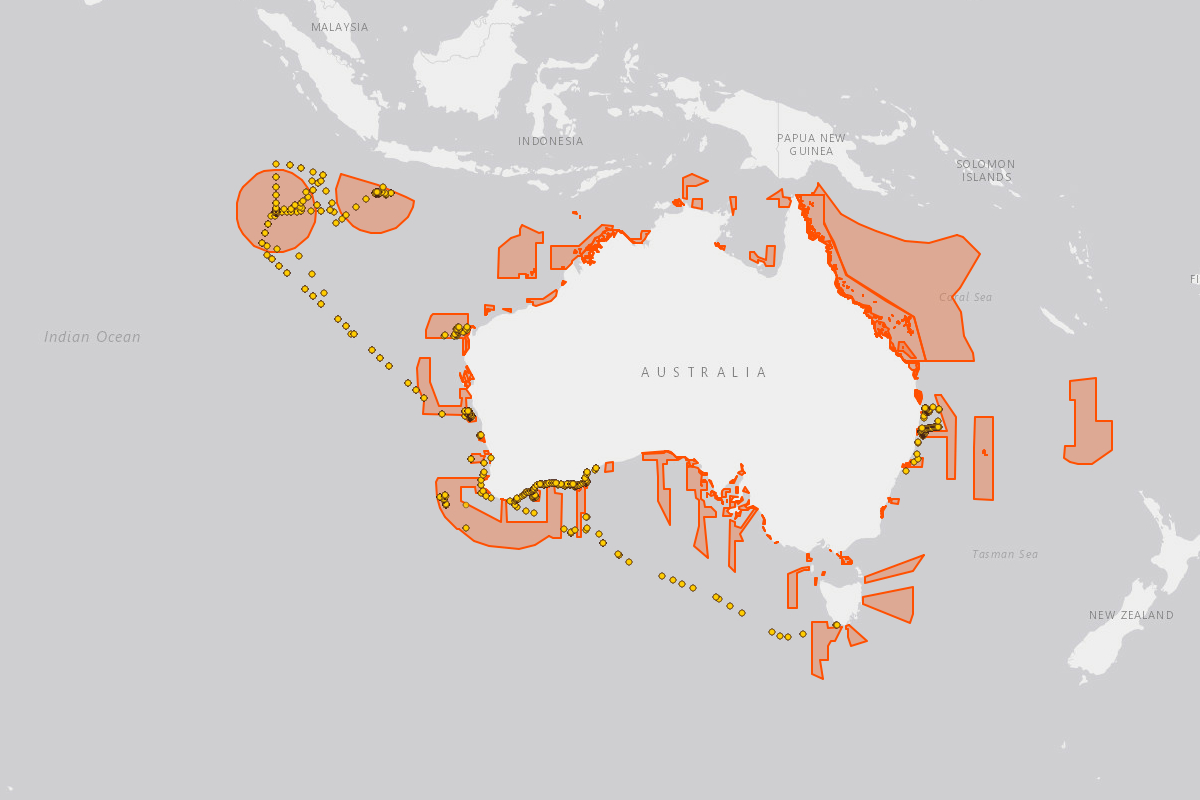
The Christmas Island Marine Park is known to harbor 10 species of sharks: whale shark (Rhincodon typus), silvertip shark (Carcharhinus albimarginatus), grey reef shark (Carcharhinus amblyrhynchos), silky shark (Carcharhinus falciformis), oceanic whitetip shark (Carcharhinus longimanus), tiger shark (Galeocerdo cuvier), whitetip reef shark (Triaenodon obesus), scalloped hammerhead shark (Sphyrna lewini), great hammerhead shark (Sphyrna mokarran), and pelagic thresher shark (Alopias pelagicus).[1]
From a single expedition that sampled 29 sites around Christmas Island in 2023, eDNA approaches were able to detect four of these species gray reef shark (Carcharhinus amblyrhynchos), tiger shark (Galeocerdo cuvier), whitetip reef shark (Triaenodon obesus), and scalloped hammerhead shark (Sphyrna lewini), as well as two ray species, blotched fantail ray (Taeniurops meyeni) and Chilean devil ray (Mobula tarapacana).
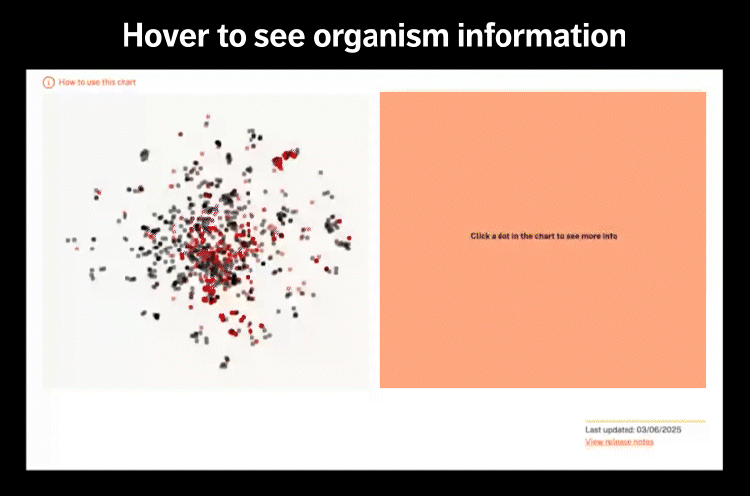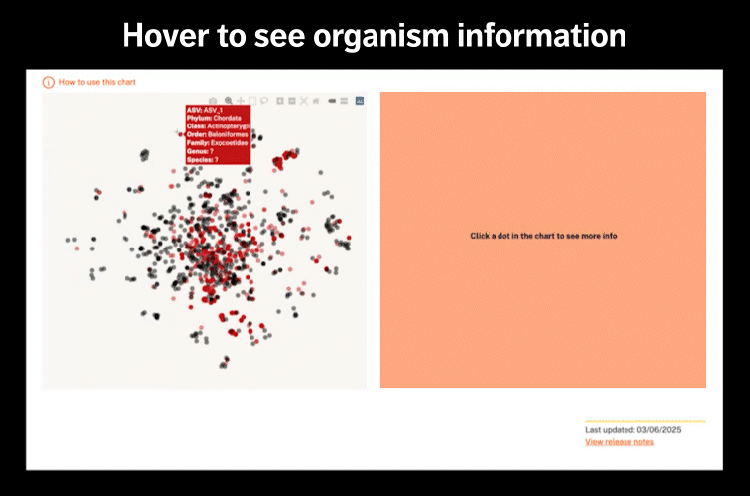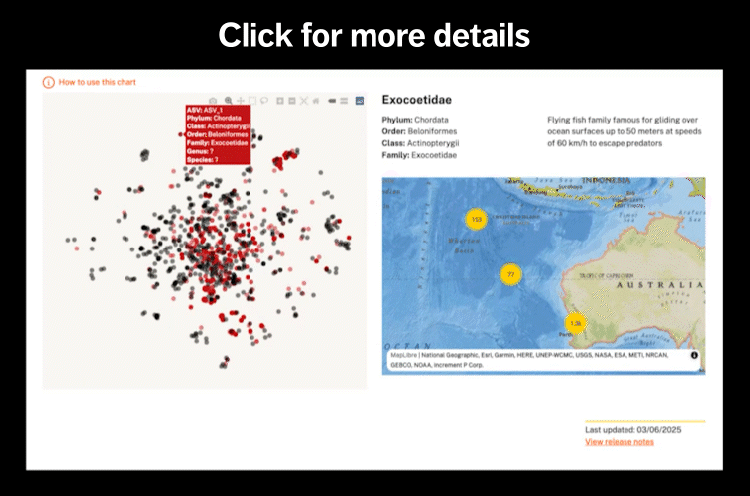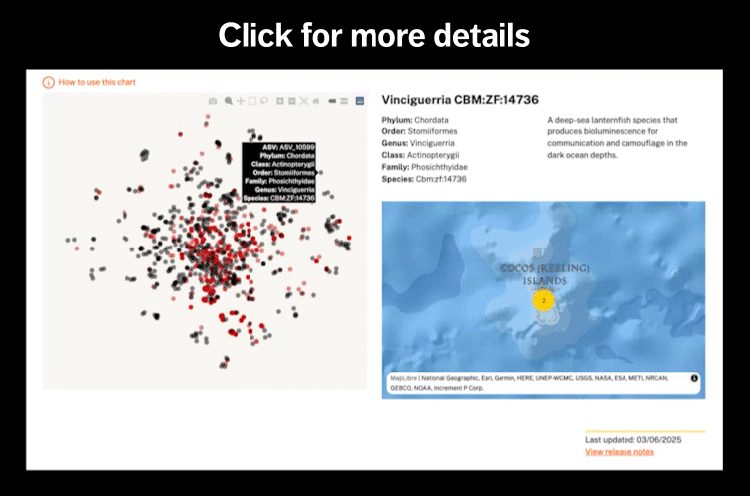
Local fisheries on Christmas Island rely on large pelagic species like wahoo (Acanthocybium solandri, tengiri), yellowfin tuna (Thunnus albacares, tuna sirip kuning), dogtooth tuna (Gymnosarda unicolor, haruan tasik), and bluefin trevally (Caranx melampygus, ikan puteh biru. Wahoo was detected at 17 sites, tuna species were detected across 5 sites, and trevallies and jacks were detected across 23 sites.
Additionally, the deeper waters surrounding Christmas Island are unique habitats for valued snapper and jobfish fisheries species including midnight snapper (Macolor macularis, ikan tau song), ruby snapper (Etelis carbunculus, salman merah) and green jobfish (Aprion virescens, salman biru) which we were collectively identified across 19 sites.
These results highlight the power of eDNA – in just one expedition eDNA identified many of the known shark species and several fish species that are important to the local community, paving the way for more detailed analyses and species-level monitoring.